This post is a part of AI April initiative, where each day of April my colleagues publish new original article related to AI, Machine Learning and Microsoft. Have a look at the Calendar to find other interesting articles that have already been published, and keep checking that page during the month.
In the last couple of years, we have witnessed significant progress in many tasks related to natural language processing, mostly thanks to transformer models and BERT architecture. In general, BERT can be effectively used for many tasks, including text classification, named entity extraction, prediction of masked words in context, and even question answering.
Training BERT model from scratch is very resource-intensive, and most of the applications rely on pre-trained models, using them for feature extraction, or for some gentle fine-tuning. So the original language-processing task that we need to solve can be decomposed into a pipeline of more simple steps, BERT feature extraction being one of them, and others being tokenization, applying TF-IDF ranking to a set of documents, or just plain classification.
This pipeline can be viewed as a set of processing steps represented by some neural networks. Taking text classification as an example, we will have BERT preprocessor step that extracts features, followed by a classification step. This series of steps can be composed into one neural architecture, and trained end-to-end on our text dataset.
DeepPavlov
Here comes DeepPavlov library. It mainly does those things for you:
- Allows you to declaratively describe text processing pipeline as a series of steps by writing a config.
- Provides a number of pre-defined steps, including pre-trained models such as BERT preprocessor
- Provides a number of pre-defined configs that you can use to solve common tasks
- Perform pipepline training and inference from Python SDK or from command-line interface
The library actually does much more than that, giving you the ability to run it in the REST API mode, or as chatbot backend to [Microsoft Bot Framework][BotFramework]. However, we will focus on core functionality that makes DeepPavlov as useful for natural language processing, as Keras is for images.
You can easily explore DeepPavlov functionality by playing with an interactive web demo at https://demo.deeppavlov.ai.
BERT Classification with DeepPavlov
Consider again the problem of text classification using BERT embeddings. DeepPavlov contains a number of pre-defined config files for that, for example, look at Twitter Sentiment Classification. In this config, chainer section describes the pipeline, which consists of the following steps:
simple_vocab is used to convert expected output (y), which is a class name, into numeric id (y_ids)transformers_bert_preprocessor takes the input text x, and produces a set of data which BERT network expectstransformers_bert_embedder actually produces BERT embeddings for input textone_hotter encodes y_ids into one-hot encoding, needed by the final layer of the classifierkeras_classification_model is the classification model, which is multi-layer CNN with defined parametersproba2labels final layer converts the output of the network to corresponding label
Config also defines the dataset_reader to describe the format and path to input data, and training parameters in train section, as well as some other less important stuff.
Once we have defined the config, we can train it from a command-line like this:
python -m deeppavlov install sentiment_twitter_bert_emb.json
python -m deeppavlov download sentiment_twitter_bert_emb.json
python -m deeppavlov train sentiment_twitter_bert_emb.json
install command installs all required libraries to perform the process (such as Keras, transformers, etc.), the second line downloads all pre-trained models, and the last line performs the training.
Once the model has been trained, we can interact with it from the command-line:
python -m deeppavlov interact sentiment_twitter_bert_emb.json
And we can also use the model from any Python code:
model = build_model(configs.classifiers.sentiment_twitter_bert_emb)
result = model(["This is input tweet that I want to analyze"])
Open Domain Question Answering
One of the most interesting tasks that can be performed using BERT is called Open Domain Question Answering, or ODQA for short. We want to be able to give a computer a bunch of text to read, and expect it to give specific answers to general questions. The problem of Open Domain question answering refers to the fact that we do not refer to one document, but rather to a very broad domain of knowledge.
ODQA typically works in two stages:
- First, we try to find the best matching document for the query, performing the task of information retrieval.
- Then, we process the document by a network to produce the specific answer from that document (machine comprehension).
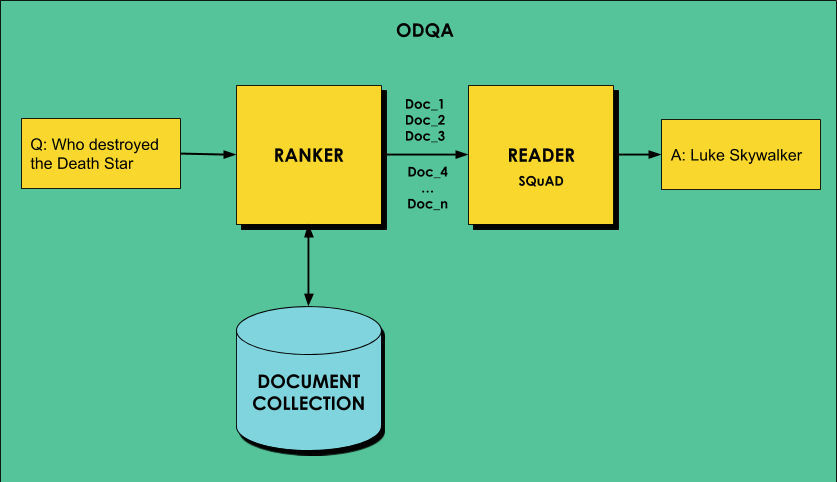
The process of using DeepPavlov for ODQA has been described quite well in this blog post, however, they are using R-NET for the second stage, and not BERT. In this post, we will apply ODQA with BERT to the COVID-19 Open Research Dataset, which contains more than 52,000 scholarly articles on COVID-19.
Getting Training Data with Azure ML
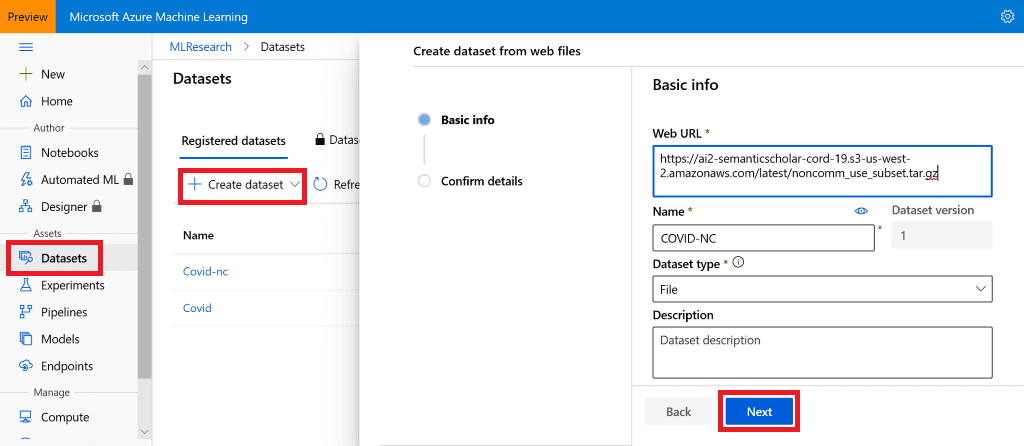
For training, I will use Azure Machine Learning, in particular Notebooks. The simplest way to get data into AzureML is to create a Dataset. You can see all available data at Semantic Scholar Page. We will use non-commercial subset, located here.
To define a dataset, I will use Azure ML Portal, and create a dataset from web files, chosing file as the dataset type. The only thing I need to do is to provide the URL.
To access the dataset, I will need a notebook and compute. Because the ODQA task is quite intensive, and a lot of memory is needed, I will create a big compute with NC12 virtual machine with 112Gb or RAM. The process of creating a compute and a notebook is described here.
To access the dataset from my notebook, I will need the following code:
from azureml.core import Workspace, Dataset
workspace = Workspace.from_config()
dataset = Dataset.get_by_name(workspace, name='COVID-NC')
The dataset contains one compressed .tar.gz-file. To decompress it, I will mount the dataset as a directory, and perform UNIX command:
mnt_ctx = dataset.mount('data')
mnt_ctx.start()
!tar -xvzf ./data/noncomm_use_subset.tar.gz
mnt_ctx.stop()
All text is contained within noncomm_use_subset directory as .json files, which contain abstract and full paper text in abstract and body_text fields. To extract just the text into separate text files, I will write short Python code:
from os.path import basename
def get_text(s):
return ' '.join([x['text'] for x in s])
os.makedirs('text',exist_ok=True)
for fn in glob.glob('noncomm_use_subset/pdf_json/*'):
with open(fn) as f:
x = json.load(f)
nfn = os.path.join('text',basename(fn).replace('.json','.txt'))
with open(nfn,'w') as f:
f.write(get_text(x['abstract']))
f.write(get_text(x['body_text']))
Now we will have a directory called text, with all papers in the textual form. We can get rid of the original directory:
!rm -fr noncomm_use_subset
Setting up ODQA model
First of all, let’s set up original pre-trained ODQA model in DeepPavlov. There is an existing config named en_odqa_infer_wiki that we can use:
import sys
!{sys.executable} -m pip --quiet install deeppavlov
!{sys.executable} -m deeppavlov install en_odqa_infer_wiki
!{sys.executable} -m deeppavlov download en_odqa_infer_wiki
This will take quite some time to download, and you will have time to realize how lucky you are to be using cloud resources, and not your own computer. Downloading cloud-to-cloud is much faster!
To use this model from Python code, we just need to build the model from config, and apply it to the text:
from deeppavlov import configs
from deeppavlov.core.commands.infer import build_model
odqa = build_model(configs.odqa.en_odqa_infer_wiki)
answers = odqa([ "Where did guinea pigs originate?",
"When did the Lynmouth floods happen?" ])
The answers we will get:
['Andes of South America', '1804']
If we try to ask the network something about coronavirus, here are the answers we will get:
- What is coronavirus? – a strain of a particular virus
- What is COVID-19? – nest on roofs or in church towers
- Where did COVID-19 originate? – northern coast of Appat
- When was the last pandemic? – 1968
Far from perfect! Those answers come from the old Wikipedia text, which the model has been trained on. Now we need to re-train the document extractor on our own data.
Training the model
The process of training ranker on your own data is described in DeepPavlov Blog. Because ODQA model uses en_ranker_tfidf_wiki as a ranker, we can load its config separately, and substitute data_path that is uses for training.
from deeppavlov.core.common.file import read_json
model_config = read_json(configs.doc_retrieval.en_ranker_tfidf_wiki)
model_config["dataset_reader"]["data_path"] = os.path.join(os.getcwd(),"text")
model_config["dataset_reader"]["dataset_format"] = "txt"
model_config["train"]["batch_size"] = 1000
We also decrease the batch size here, otherwise the training process will not fit into memory. Again time to realize that we probably do not have a physical machine at hand with 112 Gb of RAM.
Now let’s train the model and see how it performs:
doc_retrieval = train_model(model_config)
doc_retrieval(['hydroxychloroquine'])
This should give us the list of file names that are relevant to the specified keyword.
Now let’s instantiate actual ODQA model and see how it performs:
# Download all the SQuAD models
squad = build_model(configs.squad.multi_squad_noans_infer, download = True)
# Do not download the ODQA models, we've just trained it
odqa = build_model(configs.odqa.en_odqa_infer_wiki, download = False)
odqa(["what is coronavirus?","is hydroxychloroquine suitable?"])
The answers we will get:
['an imperfect gold standard for identifying King County influenza admissions',
'viral hepatitis']
Still not so good…
Using BERT for question answering
DeepPavlov has two pre-trained models for Question Answering, trained on Stanford Question Answering Dataset (SQuAD): R-NET and BERT. In the previous example, R-NET was used. We will now switch it to BERT. squad_bert_infer config is the good starting point for using BERT Q&A inference:
!{sys.executable} -m deeppavlov install squad_bert_infer
bsquad = build_model(configs.squad.squad_bert_infer, download = True)
If you look at ODQA config, the following part is responsible for question answering:
{
"class_name": "logit_ranker",
"squad_model":
{"config_path": ".../multi_squad_noans_infer.json"},
"in": ["chunks","questions"],
"out": ["best_answer","best_answer_score"]
}
To change the question answering engine in the overall ODQA model, we need to substitute the squad_model field in the config:
odqa_config = read_json(configs.odqa.en_odqa_infer_wiki)
odqa_config['chainer']['pipe'][-1]['squad_model']['config_path'] =
'{CONFIGS_PATH}/squad/squad_bert_infer.json'
Now we build and use the model in the same way as did before:
odqa = build_model(odqa_config, download = False)
odqa(["what is coronavirus?",
"is hydroxychloroquine suitable?",
"which drugs should be used?"])
Below are some questions and answers we were able to get from the resulting model:
| Question | Answer |
|---|
| what is coronavirus? | respiratory tract infection |
| is hydroxychloroquine suitable? | well tolerated |
| which drugs should be used? | antibiotics, lactulose, probiotics |
| what is incubation period? | 3-5 days |
| is patient infectious during incubation period? | MERS is not contagious |
| how to contaminate virus? | helper-cell-based rescue system cells |
| what is coronavirus type? | enveloped single stranded RNA viruses |
| what are covid symptoms? | insomnia, poor appetite, fatigue, and attention deficit |
| what is reproductive number? | 5.2 |
| what is the lethality? | 10% |
| where did covid-19 originate? | uveal melanocytes |
| is antibiotics therapy effective? | less effective |
| what are effective drugs? | M2, neuraminidase, polymerase, attachment and signal-transduction inhibitors |
| what is effective against covid? | Neuraminidase inhibitors |
| is covid similar to sars? | All coronaviruses share a very similar organization in their functional and structural genes |
| what is covid similar to? | thrombogenesis |
Conclusion
The main goal of this post was to demonstrate how to use Azure Machine Learning together with DeepPavlov NLP library to do something cool. I did not have the goad to make some unexpected findings in the COVID dataset – ODQA approach is probably not the best way to do it. However, DeepPavlov can be used in the similar manner to perform other tasks on this dataset, for example entity extraction can be used to cluster papers into thematic groups, or to index them based on entities. I definitely encourage readers to check out the COVID challenge on Kaggle, and see if you can come up with original ideas that can be implemented using DeepPavlov and Azure Machine Learning.
Azure ML infrastructure and DeepPavlov library helped me to get the experiment I described running in a few hours. Taking this example as a starting point, you can achieve much better results. If you do - please share them with the community! Data Science can do wonderful things, but even more so when many people start working on a problem together!